diaGrabber.target¶
Matrix¶

Baseclass for all matrices¶
- class diaGrabber.target._matrix.matrixBase(sourceClassList)¶
Bases: diaGrabber.target._matrixMethods.matrixMethods
Base-Class for all matix-like-targets. Includes every method that is equal for them.
Parameters: sourceClassList (tuple, list) – One or more instances of source-classes Using more source-classes can be usefull to:
- combine (same merge-dimensions)
- compare (different merge-dimensions)
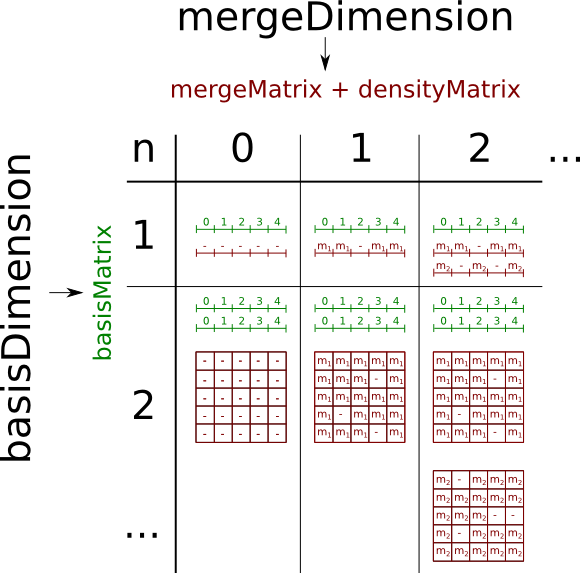
values. When more sources are used each source has to have the same basis (with same names and units) Different ranges and resolutions will be fitted. If different sources includes mergeDimensions of the same name (and unit) their values are assembled. Depending on the number of used basis- and mergeDimensions all matrix-targets create three different matrices:
- basisMatrix (1D-numpy.array)
- mergeMatrix (nD-numpy.array)
- densityMatrix (nD-numpy.array)
...
- fill()¶
Readout the whole source, then continue.
- fillInteractive(plotInstance, **kwargs)¶
Fill the matrix with the source-values and plot the progress. Therefore a plotInstance which is able to plot interactive is needed. kwargs can be those from the plot-method of the used ‘plotInstance’
- load(name, folder='', ftype='bin')¶
Load previous saved matrices. Only ftype=’bin’ is supported at the moment.
- save(**kwargs)¶
- Save the following matrices (of all mergeDimensions) to file:
- basisMatrix
- mergeMatrix
- densityMatrix
Required kwargs (“keyword arguments”) are:
Keyword Type Example Description name string “test” prename of the saved files. Optional kwargs (“keyword arguments”) are:
Keyword Type Default Description folder string “” Name of the folder to save in ftype string “bin” “bin”: output is saved in computer-readable binary-form, “txt”: output is saved in a human-readable-form
coarseMatrix¶
- class diaGrabber.target.coarseMatrix.coarseMatrix(sourceClassList)[source]¶
Bases: diaGrabber.target._matrix.matrixBase
Takes one or more source-instances as list or tuple. Create like this:
myMatrix = diaGrabber.target.coarseMatrix( (mySource1,...mySourceN) )
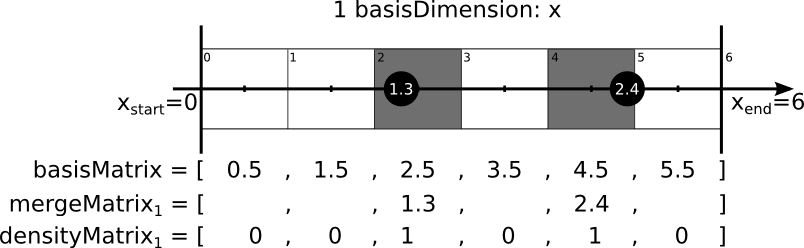
Coarse matrices store values of the mergeDimension at the nearest point in the mergeMatrix given by the difference of the values of the basisDimension and and the values of the basisMatrix. The following images iluustrated this procedure.

fineMatrix¶
- class diaGrabber.target.fineMatrix.fineMatrix(sourceClassList)[source]¶
Bases: diaGrabber.target._matrix.matrixBase
Takes one or more source-instances as list. Create like this:
myMatrix = diaGrabber.target.fineMatrix( (mySource1,...mySourceN) )
This class isn’t as fast as coarseMatrix but can assign values in a more accurate way if the following condition is fullfilled:
Note
number of datapoints in matrix>> matrix-resolution
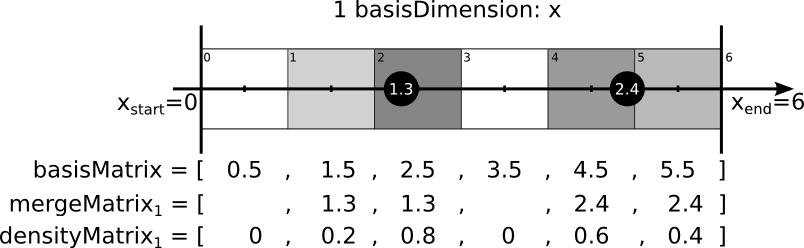
In a fineMatrix the values of the mergeDimensions were stored in the mergeMatrix at the nearest point (analoque to the procedure of the coarseMatrix) AND all points near this point. Depending on the number of basisDimensions the following number of positions were filled:
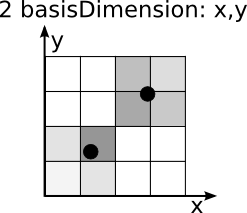
nBasis nPositions 1 2 2 4 3 8 This splitted values are wheighted through its values in the densityMatrix - ther pointsdensity. The sum of this pointsdensity is allways one. The following images should illustrate this procedure:
Methods to process filled matrices¶
- class diaGrabber.target._matrix.matrixMethods¶
This class includes a list of methods inherited (and hence callable) by all matrix-like targets.
- autoZoom(**kwargs)¶
Required kwargs (“keyword arguments”) are:
Keyword Type Example Description mergeName str myMergeName the name of the merge-dim to do the method on value float/string max The merge-value to zoom in. Type ‘min’ or ‘max’ to use the equivalent extrem-value in the matrix scale string ‘relative’ OR ‘absolute’ level float 0.3 The relative zoom-level 0.3 means +-30% around the zoom-point Optional kwargs (“keyword arguments”) are:
Keyword Type Default Description basisNames list(basisNames) [{all}] Which basisDimensions get a new scale. operator string “==” The zoom-point is defined as the first point in matrix where a value in “==” (equal), “>” (bigger) etc. than the given value
- copyMerge(presentMergeName, newMergeName)¶
copy an existent mergeDim, including its merge- and densityMatrix. Return the copied mergeDim
- interpolateFast(**kwargs)¶
interpolate NaNs (empty points) in matrix using scipy.interpolate.griddata.
Optional kwargs (“keyword arguments”) are:
Keyword Type Default Description mergeName list(mergeDim) [{all}] one or more merge-dims to do the method on method str ‘nearest’ {“nearest”, “linear”, “cubic”}
- interpolateFine(**kwargs)¶
interpolate NaNs (empty points) in matrix. In contrast to interpolateFast() this function is comparatively slow. Therefore it can take a some minutes to finish interpolation on mergeMatrices that have reslutions > 30 in each basisDimension. However this function enables you to:
- blurring between points (focusExponent). Thus it is possible to get smooth intersections between heavy scattered values.
- extrapolate
- limit the maximum interpolation/extrapolation distance (related to the unit of the chosen basisDimension)
- weight distances in each basisDimension
Optional kwargs (“keyword arguments”) are:
Keyword Type Default Description mergeName list(mergeDim) [{all}] one or more merge-dims to do the method on focusExponent float 10 blurring between points (sharpness will increase with increasing focusExponent) evalPointDensity bool True weights the moments: ‘moment = pointDensity*moment’ maxDistance dict {None} {basisName:maxDistance,..} distanceFactor dict {1} {basisName:factor,..} \[moment = 1/distance^{focusExponent}\]
- posterize(**kwargs)¶
This method discretize/posterize the values in the mergeMatrix (of a given mergeDimension) to a given ammount of values (e.g. nValues= 4 ) or to a given list or values (e.g. values = [1,2,4])
Optional kwargs (“keyword arguments”) are:
Keyword Type Default Description mergeName list(mergeNames) [{all}] one or more merge-dims to do the method on nValues int 5 the amount of different values values list None given different values
- transformDim(**kwargs)¶
Keyword Type Default Description mergeName list(mergeDim) [{all}] one or more merge-dims to do the method on rebuildMatrix bool False ‘True’ does: transformMatrixToPoints() _buildMatrices() fill()
- transformMatrixToPoints(givenMultiDimMeshgrid=None, mergeIndex=0)¶
transform all not-empty parts of a matrix to discrete points
Returns: list(list(basis-values)), list(merge-values)
- transformPointsToMatrix(points, merge, mergeIndex=0)¶
Counter piece of transformMatrixToPoints()
